- Omics” refers to a collective group of technologies that have been helpful in understanding biology at the molecular level by analysing data with systems biology and bioinformatic methods.
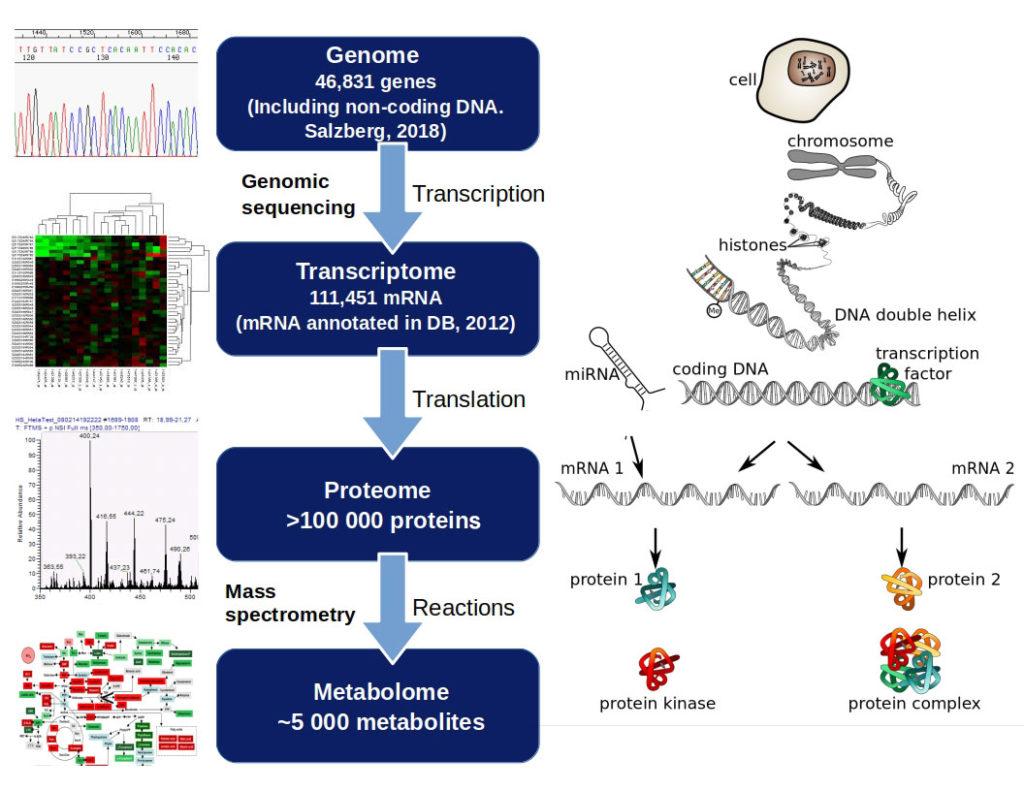
- “Omics” experiments are conducted in high-throughput assays, including genomics, proteomics, metabolomics, transcriptomics, epigenomics, glycomics, lipomics and microbiomics. Such assays target different regulatory levels in the cell (Figure 1).
- The basis of these approaches is that a complex system can be better understood when analysed as a whole.
- Merging of different levels of analysis approached by “omics” technologies is still in progress.
- For any “omics” experiment, the initial design is very important and may consider the use of suitable biological samples, the type of the assay, sample size, the technical/biological variation, validation methods and the type of algorithms that will be used for its analysis.
Genomics
- Genomics was the first “omics” technology to be developed and has resulted in massive amounts of sequencing data requiring high computational capacity and new analysing algorithms.
- Genomics is the systematic study of the total DNA within an organism or cell.
- The human genome contains 3.2 billion bases and estimated 30 000–40 000 protein‐coding genes.
- Genomic and transcriptomic research have progressed due to advances in microarray technologies.
- DNA microarrays can simultaneously measure the expression of thousands of genes.
- They can reveal abnormalities such as chromosomal insertions, deletions, abnormal chromosomal numbers (comparative genomic hybridization), single nucleotide polymorphisms (SNPs) and more.
- Genomics has progressed beyond sequencing of organisms (structural genomics) to identifying the function of the encoded genes (functional genomics).
- Gene annotation includes obtaining the greatest amount of information for a genetic sequence such as: encoded protein, amino acid sequence, chromosomal location, identification of start/stop codons or promoter regions.
- The multiple applications of genomics have resulted in more specific areas, like pharmacogenomics which elucidates the role of inheritance in drug response, among others. Pharmacogenomics is especially important for emerging therapies focused on personalised medicine.
Transcriptomics
- The transcriptome is the total mRNA (template for protein synthesis) in a cell or organism.
- The transcriptome reflects genes that are actively expressed at any given moment, this is highly dependent on external stimuli.
- For transcriptome microarrays, the RNA is extracted from the organism/cell of study, processed by reverse transcription and stained with fluorescent dyes to obtain labelled control/case complementary DNA (cDNA).
- The cDNA probe is then amplified by polymerase chain reaction (PCR) and hybridized with a microarray slide.
- An ultraviolet laser is used to scan the slide which detects the amount of fluorescent signal for each gene.
- One limitation of gene expression microarrays is that they measure changes in mRNA abundance, which is in turn susceptible to many types of regulatory mechanisms before protein production.
- Thus, modifications in mRNA quantification can’t be directly interpreted as a functional effect.
Proteomics
- Proteins are the primary structural and functional molecules in the cell, they are made up of a linear arrangements of amino acids.
- The linear polypeptide chains are folded into secondary and tertiary structures to form a functional protein.
- The proteome is defined as the set of all expressed proteins in a determined sample (cell, tissue or organism).
- Thanks to the development of bioinformatic tools, proteomics data can be matched with libraries to identify protein pathways and networks with the aim of understanding the functional relevance of protein changes.
- The proteome is a dynamic reflection of both, genes and the micro/macro environment, and is frequently studied with the aim of discovering novel biomarkers for diseases.
- Mass spectrometry (MS) is the most common method used in proteomic and metabolomic research.
- MS is an analytical technique that measures the mass‐to‐charge (m/z) ratio of charged particles.
- In MS the sample is ionized from neutral proteins, peptides or metabolites.
- The ions are then separated according to their mass‐to‐charge ratio and detected to create a mass spectrum.
- A mass spectrum contains characteristic signals of molecular mass that, when compared to MS libraries, allow the identification of the compounds presented in the sample.
- Ionization techniques have improved MS resolution. Most recurrent techniques include electrospray ionization (ESI) and matrix‐assisted laser desorption/ionization (MALDI).
- Major considerations for a proteomic experiment involve protein concentration, purification and digestion.
- Purification of protein samples has improved with chromatography techniques that reduce the complexity of the target fluid.
- Sample processing depends on the biological sample. For example, urine provides different analytical challenges to plasma, including the need for ultrafiltration and precipitation to remove salts.
Metabolomics
- Metabolomics refers to the analysis of biochemical signals (metabolites) reflecting the metabolic profile of a cell, tissue or organism under a given set of conditions.
- The metabolome is the final downstream product of gene transcription and reflects the amplified relative changes that may be occurring at a transcriptional and protein level.
- Metabolomic analysis involves the analysis of many different low-molecular-weight compounds, making it more physically and chemically complex than the other ‘omic’ assays.
- Beside the biochemical products of metabolic pathways, the metabolome also considers metabolites taken in from external environments or symbiotic relationships.
- As mentioned for proteomics, metabolomic samples also need to be fractionated (by chromatography or electrophoresis).
- The fractionation steps use the different chemical/physical properties of the biomolecules in the sample and enables the separation of proteins/peptides/metabolites in liquid or gas phase.
- Some analytical platforms use nuclear magnetic resonance (NMR) spectroscopy and infra‐red spectroscopy. However, NMR-spectroscopy has the disadvantage of poor sensitivity.
Applied omics: Immunomics
- All genes and proteins taking part in immune responses are referred to as “immunome” and it excludes genes and proteins that are expressed in cell types other than in immune cells.
- All immune reactions activated as consequence of a host/antigen interaction are referred to as “immunome reactions”.
- Immunomics has been applied to the study of autoimmune diseases, allergy prediction, T and B cell epitope mapping, reverse vaccination and more.
- Among the microarrays used in immunomics, there are dissociable antibody microarray, serum microarray, serological analysis of cDNA expression library (SEREX), peptide–MHC microarray and artificial antigen-presenting chips.
- Immunomic databases may include epitope information, immune cell markers, cytokine information, analysis tools, and prediction algorithms.
- An example of an immunomics database is InnateDB (www.innatedb.ca), which has been created to understand the complete network of pathways and interactions of the innate immune system responses.
Quiz
Download Relevant Resources:
Th1Th17 polarization persists following wholecell pertussis vaccination
1 file(s) 485.46 KB